 Volver al Inicio
Volver al Inicio
 Volver al Inicio
Volver al Inicio
27 de Febrero del 2023 | Jhonatan Montilla
Pandas es una librería de Python especializada en el manejo y análisis de estructuras de datos. En este artículo encontrarás la guía más completa de sus funciones para la manipulación de datos.
| 1. Describe | 7. Groupby | 13. Fillna | 19. Value Counts | 25. Pivot Table | 31. Rolling |
| 2. Info | 8. Select Types | 14. Interpolate | 20. Unique | 26. Loc & iLoc | 32. Transpose |
| 3. Apply | 9. Pandas Filter | 15. Drop NA | 21. Rank | 27. Assing | |
| 4. Nunique | 10. Pandas Charts | 16. Drop | 22. Query | 28. Cut | |
| 5. Sort Values | 11. Sample | 17. Replace | 23. Melt | 29. Isin | |
| 6. Rename | 12. Isnull, Isna | 18. Drop Duplicates | 24. Crosstab | 30. Value Count |
import pandas as pd
import numpy as np
import seaborn as sns
df = sns.load_dataset('penguins')
df.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
Sin parámetros se obtienen estadísticas de solo valores numéricos
df.describe()
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| count | 342.000000 | 342.000000 | 342.000000 | 342.000000 |
| mean | 43.921930 | 17.151170 | 200.915205 | 4201.754386 |
| std | 5.459584 | 1.974793 | 14.061714 | 801.954536 |
| min | 32.100000 | 13.100000 | 172.000000 | 2700.000000 |
| 25% | 39.225000 | 15.600000 | 190.000000 | 3550.000000 |
| 50% | 44.450000 | 17.300000 | 197.000000 | 4050.000000 |
| 75% | 48.500000 | 18.700000 | 213.000000 | 4750.000000 |
| max | 59.600000 | 21.500000 | 231.000000 | 6300.000000 |
df.describe(include='all')
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| count | 344 | 344 | 342.000000 | 342.000000 | 342.000000 | 342.000000 | 333 |
| unique | 3 | 3 | NaN | NaN | NaN | NaN | 2 |
| top | Adelie | Biscoe | NaN | NaN | NaN | NaN | Male |
| freq | 152 | 168 | NaN | NaN | NaN | NaN | 168 |
| mean | NaN | NaN | 43.921930 | 17.151170 | 200.915205 | 4201.754386 | NaN |
| std | NaN | NaN | 5.459584 | 1.974793 | 14.061714 | 801.954536 | NaN |
| min | NaN | NaN | 32.100000 | 13.100000 | 172.000000 | 2700.000000 | NaN |
| 25% | NaN | NaN | 39.225000 | 15.600000 | 190.000000 | 3550.000000 | NaN |
| 50% | NaN | NaN | 44.450000 | 17.300000 | 197.000000 | 4050.000000 | NaN |
| 75% | NaN | NaN | 48.500000 | 18.700000 | 213.000000 | 4750.000000 | NaN |
| max | NaN | NaN | 59.600000 | 21.500000 | 231.000000 | 6300.000000 | NaN |
df.describe(exclude='number')
| species | island | sex | |
|---|---|---|---|
| count | 344 | 344 | 333 |
| unique | 3 | 3 | 2 |
| top | Adelie | Biscoe | Male |
| freq | 152 | 168 | 168 |
df.info()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 344 entries, 0 to 343 Data columns (total 7 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 species 344 non-null object 1 island 344 non-null object 2 bill_length_mm 342 non-null float64 3 bill_depth_mm 342 non-null float64 4 flipper_length_mm 342 non-null float64 5 body_mass_g 342 non-null float64 6 sex 333 non-null object dtypes: float64(4), object(3) memory usage: 18.9+ KB
def apply_function(dataframe,col):
for i in col:
dataframe[i] = dataframe[i].apply(lambda x: x/10)
return dataframe
lis = ['bill_length_mm','bill_depth_mm','flipper_length_mm','body_mass_g']
df_mod = apply_function(df,lis)
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 3.91 | 1.87 | 18.1 | 375.0 | Male |
| 1 | Adelie | Torgersen | 3.95 | 1.74 | 18.6 | 380.0 | Female |
| 2 | Adelie | Torgersen | 4.03 | 1.80 | 19.5 | 325.0 | Female |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 3.67 | 1.93 | 19.3 | 345.0 | Female |
def nunique_function(dataframe, col):
for i in col:
print(f'{i}: {dataframe[i].nunique()}')
lis = ['species','island','sex']
nunique_function(df,lis)
species: 3 island: 3 sex: 2
def sort_values_fuction(dataframe,col,order=False):
dataframe = dataframe.sort_values(col, ascending=order)
return dataframe
df_mod = sort_values_fuction(df,'body_mass_g',True).head()
df_mod
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 190 | Chinstrap | Dream | 4.69 | 1.66 | 19.2 | 270.0 | Female |
| 64 | Adelie | Biscoe | 3.64 | 1.71 | 18.4 | 285.0 | Female |
| 58 | Adelie | Biscoe | 3.65 | 1.66 | 18.1 | 285.0 | Female |
| 116 | Adelie | Torgersen | 3.86 | 1.70 | 18.8 | 290.0 | Female |
| 98 | Adelie | Dream | 3.31 | 1.61 | 17.8 | 290.0 | Female |
df_mod = sort_values_fuction(df,['flipper_length_mm', 'body_mass_g'],True).head()
df_mod
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 28 | Adelie | Biscoe | 3.79 | 1.86 | 17.2 | 315.0 | Female |
| 20 | Adelie | Biscoe | 3.78 | 1.83 | 17.4 | 340.0 | Female |
| 122 | Adelie | Torgersen | 4.02 | 1.70 | 17.6 | 345.0 | Female |
| 98 | Adelie | Dream | 3.31 | 1.61 | 17.8 | 290.0 | Female |
| 30 | Adelie | Dream | 3.95 | 1.67 | 17.8 | 325.0 | Female |
def rename_function(dataframe,cols):
dataframe = dataframe.rename(columns=cols)
return dataframe
cols = {'species':'penguin_species'}
df_mod = rename_function(df,cols)
df_mod.head()
| penguin_species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 3.91 | 1.87 | 18.1 | 375.0 | Male |
| 1 | Adelie | Torgersen | 3.95 | 1.74 | 18.6 | 380.0 | Female |
| 2 | Adelie | Torgersen | 4.03 | 1.80 | 19.5 | 325.0 | Female |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 3.67 | 1.93 | 19.3 | 345.0 | Female |
cols = {'species':'penguin_species','island':'location'}
df_mod = rename_function(df,cols)
df_mod.head()
| penguin_species | location | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 3.91 | 1.87 | 18.1 | 375.0 | Male |
| 1 | Adelie | Torgersen | 3.95 | 1.74 | 18.6 | 380.0 | Female |
| 2 | Adelie | Torgersen | 4.03 | 1.80 | 19.5 | 325.0 | Female |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 3.67 | 1.93 | 19.3 | 345.0 | Female |
def groupby_function(dataframe,col):
dataframe = round(dataframe.groupby(col).mean(),2)
return dataframe
df_mod = groupby_function(df,'species')
df_mod
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| species | ||||
| Adelie | 3.88 | 1.83 | 19.00 | 370.07 |
| Chinstrap | 4.88 | 1.84 | 19.58 | 373.31 |
| Gentoo | 4.75 | 1.50 | 21.72 | 507.60 |
df_mod = groupby_function(df,['species', 'island'])
df_mod
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | ||
|---|---|---|---|---|---|
| species | island | ||||
| Adelie | Biscoe | 3.90 | 1.84 | 18.88 | 370.97 |
| Dream | 3.85 | 1.83 | 18.97 | 368.84 | |
| Torgersen | 3.90 | 1.84 | 19.12 | 370.64 | |
| Chinstrap | Dream | 4.88 | 1.84 | 19.58 | 373.31 |
| Gentoo | Biscoe | 4.75 | 1.50 | 21.72 | 507.60 |
def select_dtypes_function(dataframe,type):
dataframe = dataframe.select_dtypes(type)
return dataframe
df_mod = select_dtypes_function(df,'float64')
df_mod.head()
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| 0 | 3.91 | 1.87 | 18.1 | 375.0 |
| 1 | 3.95 | 1.74 | 18.6 | 380.0 |
| 2 | 4.03 | 1.80 | 19.5 | 325.0 |
| 3 | NaN | NaN | NaN | NaN |
| 4 | 3.67 | 1.93 | 19.3 | 345.0 |
def pandas_filter_funtion(dataframe,col,value,sample):
dataframe = dataframe[dataframe[col]>=value].sample(n=sample)
return dataframe
df_mod = pandas_filter_funtion(df,'bill_length_mm',40,5)
df_mod
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 295 | Gentoo | Biscoe | 48.6 | 16.0 | 230.0 | 5800.0 | Male |
| 168 | Chinstrap | Dream | 50.3 | 20.0 | 197.0 | 3300.0 | Male |
| 247 | Gentoo | Biscoe | 47.8 | 15.0 | 215.0 | 5650.0 | Male |
| 164 | Chinstrap | Dream | 47.0 | 17.3 | 185.0 | 3700.0 | Female |
| 177 | Chinstrap | Dream | 52.0 | 19.0 | 197.0 | 4150.0 | Male |
def panda_chart(dataframe,cat_var,chart_type='bar'):
chart = dataframe[cat_var].value_counts().plot(kind=chart_type)
return chart
panda_chart(df,'species')
<AxesSubplot:>
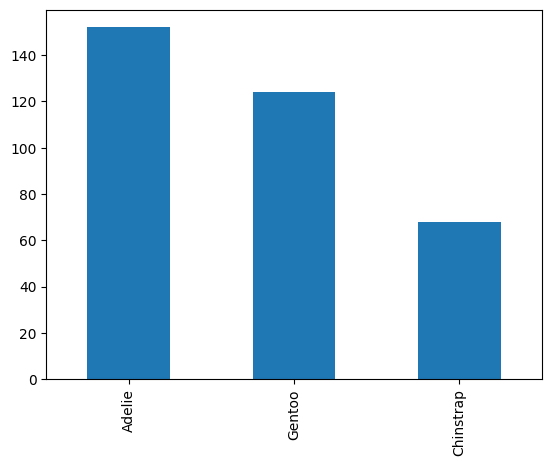
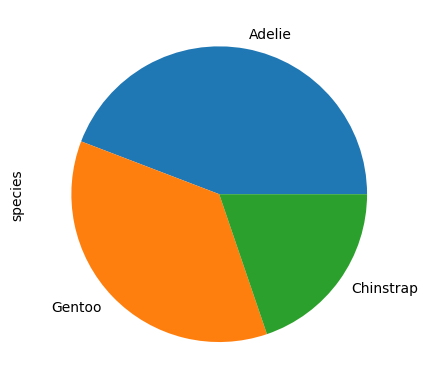
panda_chart(df,'species','pie')
<AxesSubplot:ylabel='species'>
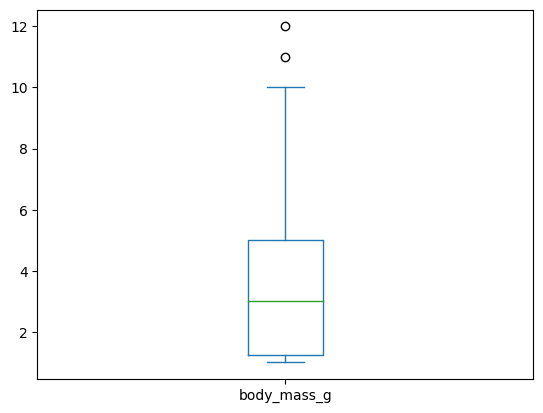
panda_chart(df,'body_mass_g','box')
<AxesSubplot:>
def sample_function(dataframe,sample,ax=0):
dataframe = dataframe.sample(n=sample, axis=ax)
return dataframe
df_mod = sample_function(df, 6) # Elige filas aleatoriamente
df_mod
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 125 | Adelie | Torgersen | 40.6 | 19.0 | 199.0 | 4000.0 | Male |
| 276 | Gentoo | Biscoe | 43.8 | 13.9 | 208.0 | 4300.0 | Female |
| 45 | Adelie | Dream | 39.6 | 18.8 | 190.0 | 4600.0 | Male |
| 83 | Adelie | Torgersen | 35.1 | 19.4 | 193.0 | 4200.0 | Male |
| 133 | Adelie | Dream | 37.5 | 18.5 | 199.0 | 4475.0 | Male |
| 214 | Chinstrap | Dream | 45.7 | 17.0 | 195.0 | 3650.0 | Female |
df_mod = sample_function(df,3,1)
df_mod.head()
| bill_depth_mm | body_mass_g | sex | |
|---|---|---|---|
| 0 | 18.7 | 3750.0 | Male |
| 1 | 17.4 | 3800.0 | Female |
| 2 | 18.0 | 3250.0 | Female |
| 3 | NaN | NaN | NaN |
| 4 | 19.3 | 3450.0 | Female |
df.isnull().sum()
species 0 island 0 bill_length_mm 2 bill_depth_mm 2 flipper_length_mm 2 body_mass_g 2 sex 11 dtype: int64
missing_values = {
'bill_length_mm':df['bill_length_mm'].median(),
'bill_depth_mm':df['bill_depth_mm'].median(),
'flipper_length_mm':df['flipper_length_mm'].median(),
'body_mass_g':df['body_mass_g'].median(),
'sex': 'Unknown'
}
df_mod = df.fillna(value=missing_values)
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.10 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.50 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.30 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | 44.45 | 17.3 | 197.0 | 4050.0 | Unknown |
| 4 | Adelie | Torgersen | 36.70 | 19.3 | 193.0 | 3450.0 | Female |
df_mod = df.fillna(method='ffill')
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
df_mod = df.fillna(method='bfill')
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
df_mod = df.interpolate()
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.70 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.40 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.00 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | 38.5 | 18.65 | 194.0 | 3350.0 | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.30 | 193.0 | 3450.0 | Female |
df_mod = df.dropna()
print('Cantidad de registros antes: ',df.shape)
print('Cantidad de registros despues: ',df_mod.shape)
Cantidad de registros antes: (344, 7) Cantidad de registros despues: (333, 7)
df_mod = df.dropna(axis=1)
print('Cantidad de registros antes: ',df.shape)
print('Cantidad de registros despues: ',df_mod.shape)
Cantidad de registros antes: (344, 7) Cantidad de registros despues: (344, 2)
df_mod = df.dropna(thresh=5) # Elimina una cantidad definido
print('Cantidad de registros antes: ',df.shape)
print('Cantidad de registros despues: ',df_mod.shape)
# df_mod.head()
Cantidad de registros antes: (344, 7) Cantidad de registros despues: (342, 7)
Esta función se usa para eliminar una cantidad de registros determinados
Elimina la primera fila del DataFrame
df_mod = df.drop(0)
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male |
df_mod = df.drop([0,1])
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male |
| 6 | Adelie | Torgersen | 38.9 | 17.8 | 181.0 | 3625.0 | Female |
df_mod = df.drop(['species', 'island'], axis=1)
df_mod.head()
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|
| 0 | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | NaN | NaN | NaN | NaN | NaN |
| 4 | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
def replace_function(dataframe,col,val,rep):
dataframe[col] = dataframe[col].replace(val,rep)
return dataframe
df_mod = replace_function(df,'species','Gentoo','Gentil')
df_mod['species'].unique()
array(['Adelie', 'Chinstrap', 'Gentil'], dtype=object)
def drop_duplicates_function(dataframe,col,kp='first',iidx=True):
dataframe = dataframe.drop_duplicates(subset=[col],keep=kp,ignore_index=iidx)
return dataframe
df_mod = drop_duplicates_function(df,'species')
df_mod
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Chinstrap | Dream | 46.5 | 17.9 | 192.0 | 3500.0 | Female |
| 2 | Gentil | Biscoe | 46.1 | 13.2 | 211.0 | 4500.0 | Female |
def value_counts_function(dataframe,col):
serie = dataframe[col].value_counts()
return serie
serie = value_counts_function(df,'species')
serie
Adelie 152 Gentil 124 Chinstrap 68 Name: species, dtype: int64
def unique_function(dataframe,col):
array = dataframe[col].unique()
return array
array = unique_function(df,'species')
array
array(['Adelie', 'Chinstrap', 'Gentil'], dtype=object)
def rank_function(dataframe,new_col,target_col,order=True):
dataframe[new_col] = dataframe[target_col].rank()
dataframe = dataframe.sort_values(new_col, ascending=order)
return dataframe
df_mod = rank_function(df,'rank','body_mass_g')
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | rank | |
|---|---|---|---|---|---|---|---|---|
| 190 | Chinstrap | Dream | 46.9 | 16.6 | 192.0 | 2700.0 | Female | 1.0 |
| 64 | Adelie | Biscoe | 36.4 | 17.1 | 184.0 | 2850.0 | Female | 2.5 |
| 58 | Adelie | Biscoe | 36.5 | 16.6 | 181.0 | 2850.0 | Female | 2.5 |
| 116 | Adelie | Torgersen | 38.6 | 17.0 | 188.0 | 2900.0 | Female | 5.5 |
| 98 | Adelie | Dream | 33.1 | 16.1 | 178.0 | 2900.0 | Female | 5.5 |
def query_function(dataframe,col_value):
dataframe = dataframe.query(col_value)
return dataframe
df_mod = query_function(df,'species == "Adelie"')
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | rank | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male | 123.0 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female | 135.5 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female | 31.0 |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female | 64.5 |
df_mod = query_function(df,'body_mass_g > 3000 and flipper_length_mm < 180')
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | rank | |
|---|---|---|---|---|---|---|---|---|
| 20 | Adelie | Biscoe | 37.8 | 18.3 | 174.0 | 3400.0 | Female | 54.5 |
| 28 | Adelie | Biscoe | 37.9 | 18.6 | 172.0 | 3150.0 | Female | 19.5 |
| 30 | Adelie | Dream | 39.5 | 16.7 | 178.0 | 3250.0 | Female | 31.0 |
| 31 | Adelie | Dream | 37.2 | 18.1 | 178.0 | 3900.0 | Male | 149.5 |
| 122 | Adelie | Torgersen | 40.2 | 17.0 | 176.0 | 3450.0 | Female | 64.5 |
def melt_function(dataframe,col,values):
dataframe = dataframe.melt(id_vars=[col], value_vars=values)
return dataframe
df_mod = melt_function(df,'species',['bill_length_mm', 'bill_depth_mm'])
df_mod.head()
| species | variable | value | |
|---|---|---|---|
| 0 | Adelie | bill_length_mm | 39.1 |
| 1 | Adelie | bill_length_mm | 39.5 |
| 2 | Adelie | bill_length_mm | 40.3 |
| 3 | Adelie | bill_length_mm | NaN |
| 4 | Adelie | bill_length_mm | 36.7 |
def crosstab_function(dataframe,col1,col2):
dataframe = pd.crosstab(dataframe[col1], dataframe[col2])
return dataframe
df_mod = crosstab_function(df,'species','sex')
df_mod
| sex | Female | Male |
|---|---|---|
| species | ||
| Adelie | 73 | 73 |
| Chinstrap | 34 | 34 |
| Gentil | 58 | 61 |
def pivot_table_function(dataframe,indx,col,values,agg_funct):
dataframe = round(dataframe.pivot_table(index=indx, columns=col, values=values,
aggfunc=agg_funct),2)
return dataframe
df_mod = pivot_table_function(df,'species','sex','bill_length_mm','mean')
df_mod
| sex | Female | Male |
|---|---|---|
| species | ||
| Adelie | 37.26 | 40.39 |
| Chinstrap | 46.57 | 51.09 |
| Gentil | 45.56 | 49.47 |
df_mod = pivot_table_function(df,['species','island'],'sex','bill_length_mm','mean')
df_mod
| sex | Female | Male | |
|---|---|---|---|
| species | island | ||
| Adelie | Biscoe | 37.36 | 40.59 |
| Dream | 36.91 | 40.07 | |
| Torgersen | 37.55 | 40.59 | |
| Chinstrap | Dream | 46.57 | 51.09 |
| Gentil | Biscoe | 45.56 | 49.47 |
Estas funciones se utilizan para seleccionar filas y columnas de un DataFrame por índice o etiqueta. La función iloc se usa para seleccionar filas y columnas mediante la indexación basada en números enteros, mientras que la función loc se usa para seleccionar filas y columnas mediante la indexación basada en etiquetas.
selecciona el primer elemento en la primera fila
df.iloc[0, 0]
'Adelie'
df.iloc[0]
species Adelie island Torgersen bill_length_mm 39.1 bill_depth_mm 18.7 flipper_length_mm 181.0 body_mass_g 3750.0 sex Male rank 123.0 Name: 0, dtype: object
df.iloc[:2]
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | rank | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male | 123.0 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female | 135.5 |
df.iloc[:, 0]
0 Adelie
1 Adelie
2 Adelie
3 Adelie
4 Adelie
...
339 Gentil
340 Gentil
341 Gentil
342 Gentil
343 Gentil
Name: species, Length: 344, dtype: object
df.iloc[:, :2]
| species | island | |
|---|---|---|
| 0 | Adelie | Torgersen |
| 1 | Adelie | Torgersen |
| 2 | Adelie | Torgersen |
| 3 | Adelie | Torgersen |
| 4 | Adelie | Torgersen |
| ... | ... | ... |
| 339 | Gentil | Biscoe |
| 340 | Gentil | Biscoe |
| 341 | Gentil | Biscoe |
| 342 | Gentil | Biscoe |
| 343 | Gentil | Biscoe |
344 rows × 2 columns
df.loc[0, "species"]
'Adelie'
df.loc[:,'sex']
0 Male
1 Female
2 Female
3 NaN
4 Female
...
339 NaN
340 Female
341 Male
342 Female
343 Male
Name: sex, Length: 344, dtype: object
df.loc[:,['species','sex']]
| species | sex | |
|---|---|---|
| 0 | Adelie | Male |
| 1 | Adelie | Female |
| 2 | Adelie | Female |
| 3 | Adelie | NaN |
| 4 | Adelie | Female |
| ... | ... | ... |
| 339 | Gentil | NaN |
| 340 | Gentil | Female |
| 341 | Gentil | Male |
| 342 | Gentil | Female |
| 343 | Gentil | Male |
344 rows × 2 columns
df_mod = df.assign(body_mass_100 = df['body_mass_g'] / 100)
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | rank | body_mass_100 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male | 123.0 | 37.5 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female | 135.5 | 38.0 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female | 31.0 | 32.5 |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female | 64.5 | 34.5 |
def cut_function(dataframe,interval_col,target_col,min,max,num,lab):
dataframe[interval_col] = pd.cut(dataframe[target_col],
bins=np.linspace(min, max, num=num),labels=lab)
return dataframe
labels = ['Botton','Low','Medium','High','Top']
df_mod = cut_function(df,'body_mass_g_binned','body_mass_g',0,6000,6,labels)
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | rank | body_mass_g_binned | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male | 123.0 | High |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female | 135.5 | High |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female | 31.0 | Medium |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female | 64.5 | Medium |
def isin_function(dataframe,col,lis):
dataframe = dataframe[dataframe[col].isin(lis)]
return dataframe
lis = ['Adelie', 'Chinstrap']
df_mod = isin_function(df,'species',lis)
df_mod
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | rank | body_mass_g_binned | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male | 123.0 | High |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female | 135.5 | High |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female | 31.0 | Medium |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female | 64.5 | Medium |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 215 | Chinstrap | Dream | 55.8 | 19.8 | 207.0 | 4000.0 | Male | 168.0 | High |
| 216 | Chinstrap | Dream | 43.5 | 18.1 | 202.0 | 3400.0 | Female | 54.5 | Medium |
| 217 | Chinstrap | Dream | 49.6 | 18.2 | 193.0 | 3775.0 | Male | 127.5 | High |
| 218 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | Male | 180.0 | High |
| 219 | Chinstrap | Dream | 50.2 | 18.7 | 198.0 | 3775.0 | Female | 127.5 | High |
220 rows × 9 columns
def value_counts_function(dataframe,col):
num = dataframe[col].value_counts()
return num
value_counts_function(df,'species')
Adelie 152 Gentil 124 Chinstrap 68 Name: species, dtype: int64
para cada ventana.
def rolling_function(dataframe,new_col,target_col,num):
dataframe[new_col] = dataframe[target_col].rolling(window=num).mean()
return dataframe
df_mod = rolling_function(df,'rolling_mean_bill_length','bill_length_mm',2)
df_mod.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | rank | body_mass_g_binned | rolling_mean_bill_length | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male | 123.0 | High | NaN |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female | 135.5 | High | 39.3 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female | 31.0 | Medium | 39.9 |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female | 64.5 | Medium | NaN |
df_mod = df.transpose() # Método abreviado df.T
df_mod.head()
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 334 | 335 | 336 | 337 | 338 | 339 | 340 | 341 | 342 | 343 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| species | Adelie | Adelie | Adelie | Adelie | Adelie | Adelie | Adelie | Adelie | Adelie | Adelie | ... | Gentil | Gentil | Gentil | Gentil | Gentil | Gentil | Gentil | Gentil | Gentil | Gentil |
| island | Torgersen | Torgersen | Torgersen | Torgersen | Torgersen | Torgersen | Torgersen | Torgersen | Torgersen | Torgersen | ... | Biscoe | Biscoe | Biscoe | Biscoe | Biscoe | Biscoe | Biscoe | Biscoe | Biscoe | Biscoe |
| bill_length_mm | 39.1 | 39.5 | 40.3 | NaN | 36.7 | 39.3 | 38.9 | 39.2 | 34.1 | 42.0 | ... | 46.2 | 55.1 | 44.5 | 48.8 | 47.2 | NaN | 46.8 | 50.4 | 45.2 | 49.9 |
| bill_depth_mm | 18.7 | 17.4 | 18.0 | NaN | 19.3 | 20.6 | 17.8 | 19.6 | 18.1 | 20.2 | ... | 14.1 | 16.0 | 15.7 | 16.2 | 13.7 | NaN | 14.3 | 15.7 | 14.8 | 16.1 |
| flipper_length_mm | 181.0 | 186.0 | 195.0 | NaN | 193.0 | 190.0 | 181.0 | 195.0 | 193.0 | 190.0 | ... | 217.0 | 230.0 | 217.0 | 222.0 | 214.0 | NaN | 215.0 | 222.0 | 212.0 | 213.0 |
5 rows × 344 columns